This post was created in July 2021 and back-dated to match the approximate date of the content. See this post or group of posts for context and more info.
Backstory
My first job out of college was working in a research lab studying Parkinson’s disease in silico. This meant automating, running, and analyzing numerous genome-wide association study (GWAS). At the time, I got curious to see if by performing multiple GWAS adjusted for known single nucleotide polymorphisms (SNPs) and identifying which SNPs change as a result of the adjustment, it might be possible to discover new cluster SNPs that could be associated with the disease. The diagrams below are my attempt to visualize this idea and explore the interactions.
Illustrations
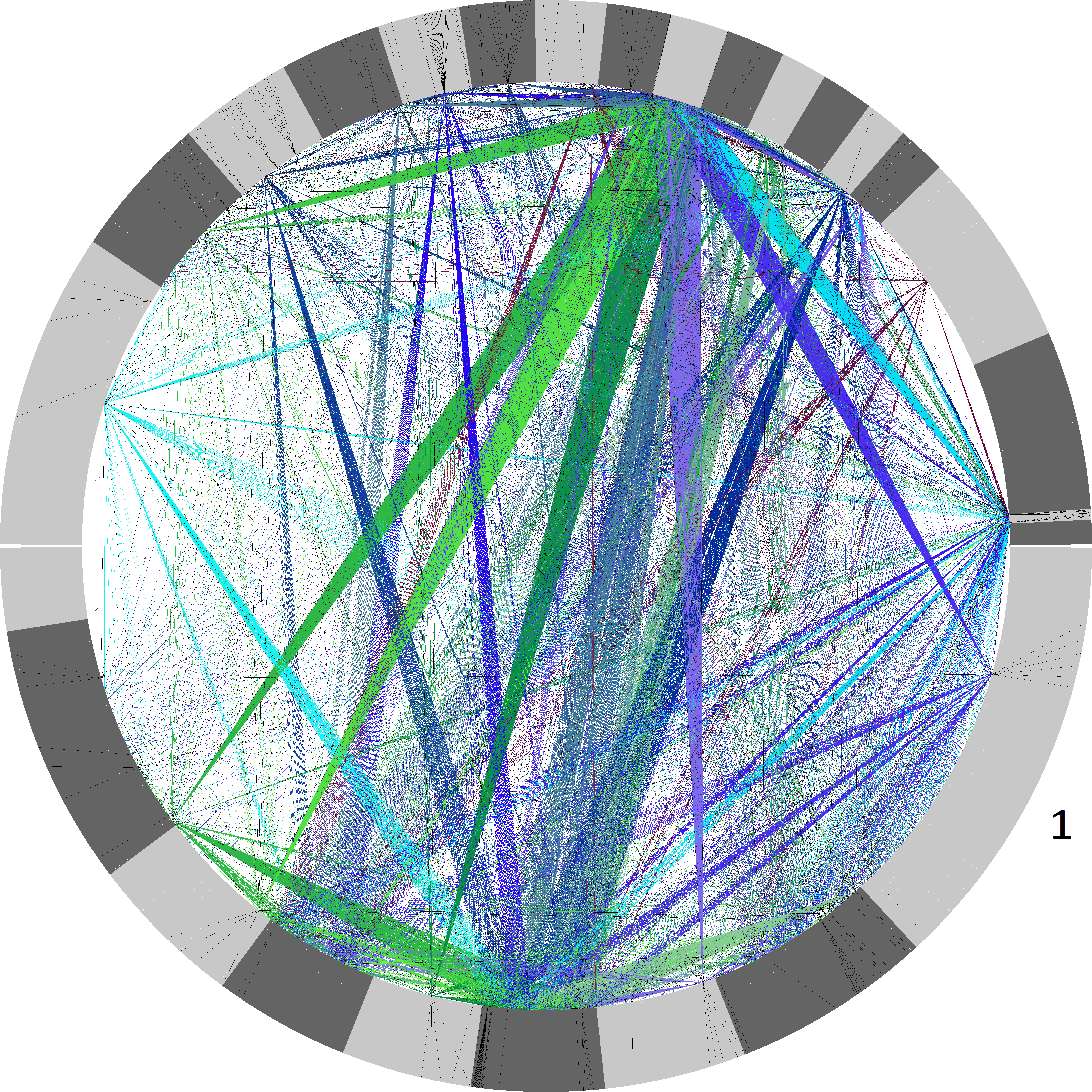 |  |
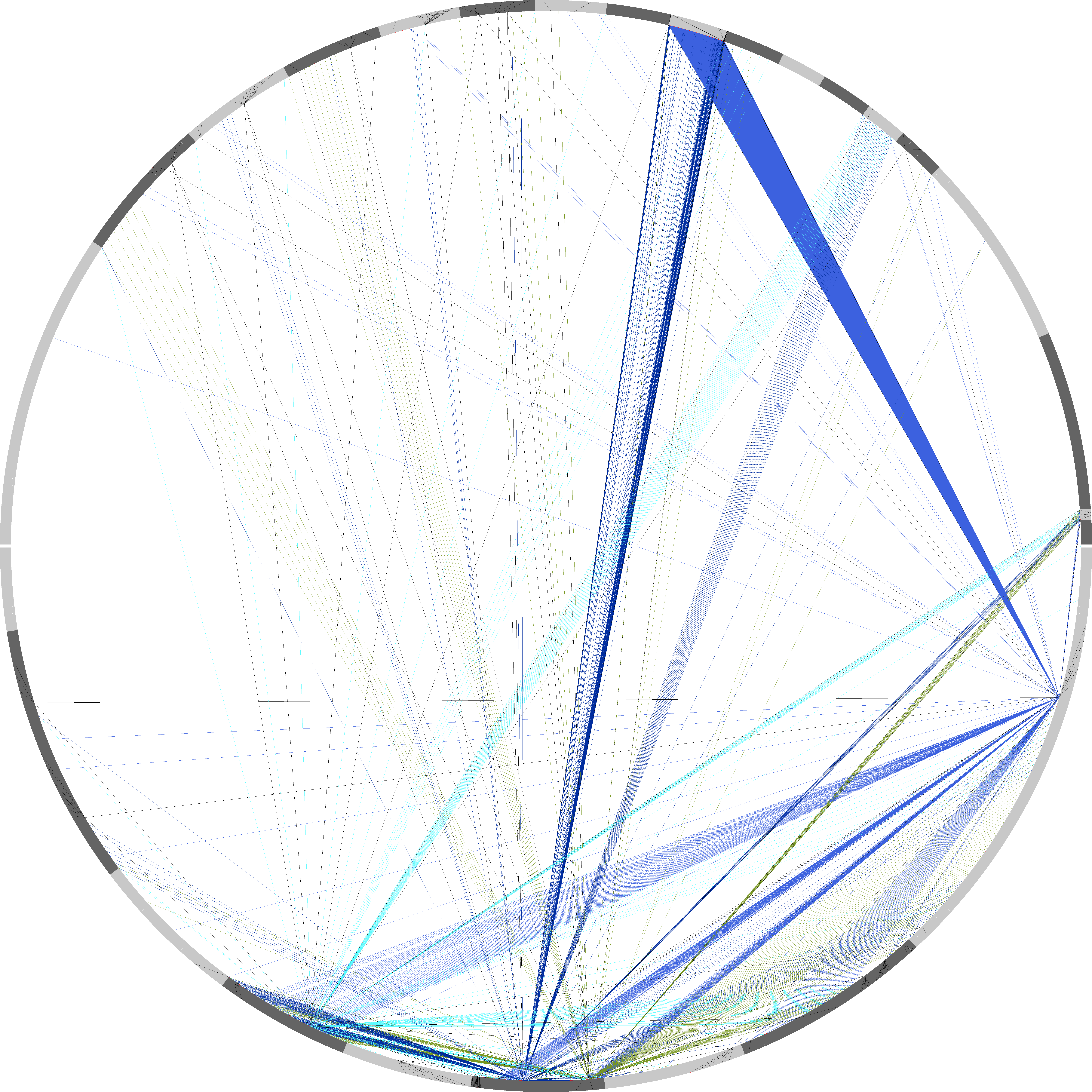 |  |
 |  |